Welcome to docking.org, software from the Shoichet and Irwin labs at UCSF!
Featured Resources
The Shoichet lab seeks to bring chemical reagents to biology, combining computation and experiment.
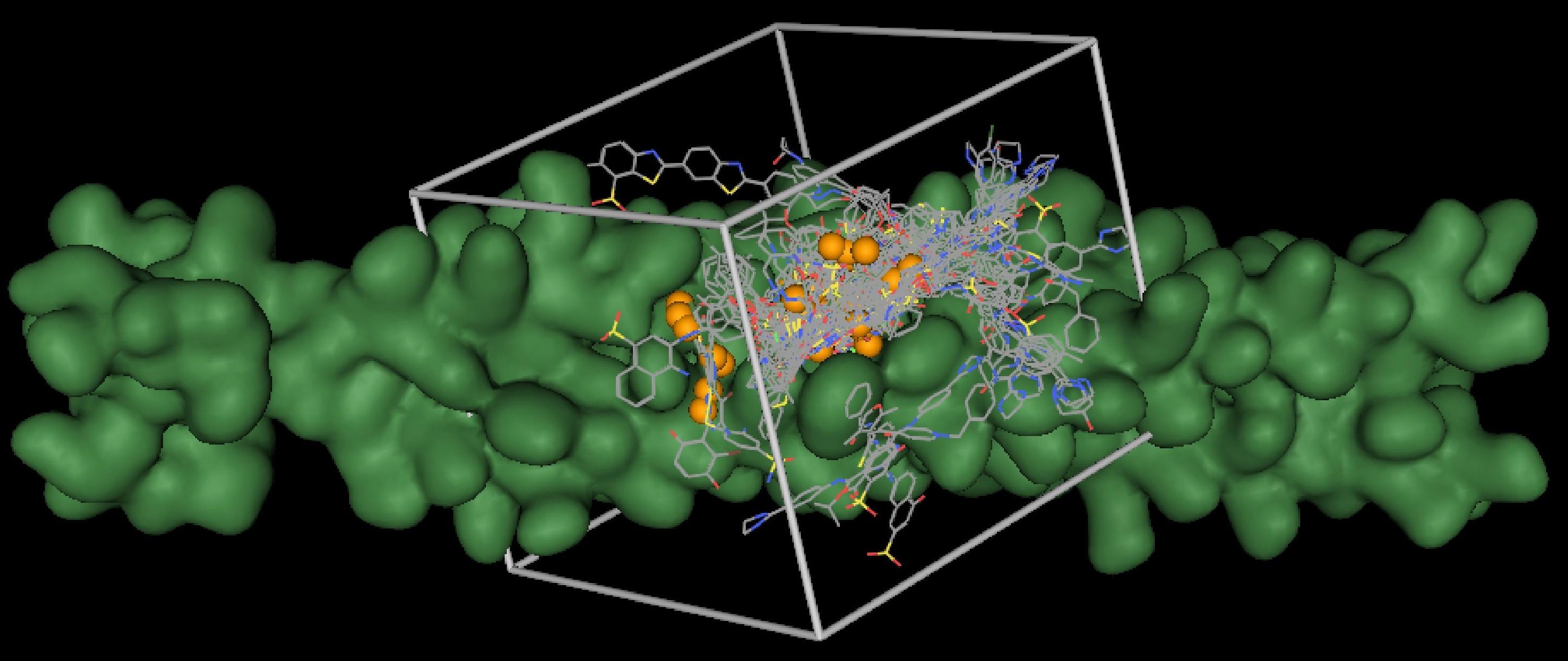
DOCK addresses the problem of "docking" molecules to each other.
All Resources
This product is part of the AWS Open Data Sponsorship Program and contains data sets that are publicly available for anyone to access and use.

Aggregator Advisor identifies molecules that are known-to aggregate or may aggregate in biochemical assays.

TA Mouse Imaging Server (AMIS), the home for mouse imaging data generated via the NIH Common fund Program for Illuminating the Druggable Genome (IDG).

NextMove Software's Arthor suite of tools aim to provide very fast substructure and similarity searching with comparatively minimal hardware infrastructure.

NextMove Software's Arthor suite of tools aim to provide very fast substructure and similarity searching with comparatively minimal hardware infrastructure.

NextMove Software's Arthor suite of tools aim to provide very fast substructure and similarity searching with comparatively minimal hardware infrastructure.

NextMove Software's Arthor suite of tools aim to provide very fast substructure and similarity searching with comparatively minimal hardware infrastructure.
Cartblanche22, an interface to ZINC-22. ZINC-22 is a free database of commercially-available compounds for virtual screening.
The Chemistry Commons connects synthetic organic chemists with biologists seeking new reagents for biology, each assisted by their medicinal and computational chemistry colleagues.

The goal of the project is to curate and disseminate information about excipients, the assumed-inactive substances found in food and drugs.

We develop software tools and databases for ligand discovery and systems pharmacology.
Large Scale Docking (LSD) refers to screening of virtual compound libraries (ZINC15, ZINC22) using the DOCK3.7/3.8 software.

The Similarity ensemble approach relates proteins based on the set-wise chemical similarity among their ligands.

SmallWorld is a chemical database indexing technology that enables rapid 2D similarity queries based upon Graph Edit Distance (GED).

SmallWorld is a chemical database indexing technology that enables rapid 2D similarity queries based upon Graph Edit Distance (GED).

SmallWorld is a chemical database indexing technology that enables rapid 2D similarity queries based upon Graph Edit Distance (GED).

SmallWorld is a chemical database indexing technology that enables rapid 2D similarity queries based upon Graph Edit Distance (GED).

A public access service for computational ligand discovery.

the wiki of the Shoichet Lab and Irwin Lab at UCSF.

A free database of commercially-available compounds for virtual screening.

A free database of commercially-available compounds for virtual screening.